Evolution and its Adversaries (Part-13)
The entire emphasis in the past by the evolutionist has been on the role of point mutations in coding regions as the primary source of evolutionary change. On the other hand, the possibility that transposable elements, or other factors, could override genetic information, was, for the most part, ignored. Now it is being said that perhaps transposition is responsible in a large part for the appearance of new species. But the proof is yet to come, writes SYED IQBAL ZAHEER.
Transposition
Although the finding of a new element affecting the genome is several decades old, its true nature and implications are just being explored seriously. And the finding is that cells re-arrange their DNA. This is known as Transposition. In the words of David S Moore, “In concept, Transposition is very simple: re-arranging coding sequences within DNA. In practice, it’s dazzlingly sophisticated …” This rearrangement is done by external agencies, which means genes (the central queen of the likes of Dawkins) cannot be considered as the master conductors. They themselves are ‘played-upon.’
Writes a scientist:
TEs (Transposable elements) now tend to be viewed as natural molecular tools that can reshape the genome … considerable evidence has accumulated suggesting that transposable elements are a reservoir of genetic innovation and that they can shape the genome, and has shown how the genome defends itself against these invaders – (Mobile Genetic Elements, Edited by Yves Bigot, 2012, epub).
Transposable elements are mobile segments of DNA that are either freely floating around inside the cell, or have been able to penetrate from outside. They are diverse in kind, and can insert themselves into the host DNA which can result in its modification and the information that it had inherited. If correctional measures are not taken by the cellular machinery, they can disrupt gene function.
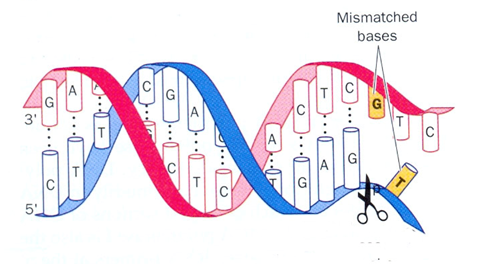
Fig. 13-1 – Interactions at genetic level. A correctional method (or editing) at the genetic level: Nucleotides G and T do not match. No protein can be made. ‘Gs’ go with ‘Cs’ alone. If left, the intended protein would not be made. Therefore, a correctional machinery arrives which cuts off the mismatching nucleotide T, and supplants the correct nucleotide viz., ‘C.’ This operation helps DNA maintain its accuracy and contradicts the idea – in a way – that genetic mutations take place despite the co-operation of genes themselves.
In further development of the process, entire stretches of DNA can be inserted by transposable elements. Gerald Karp writes:
“The first person to suggest that genetic elements were capable of moving around the genome was Barbara McClintock (in the 1950s: Au.) … After several years of careful study, she concluded that certain genetic elements were moving from one place in a chromosome to an entirely different site. She called the genetic rearrangement transposition, and the mobile genetic elements transposable elements… McClintock’s findings were largely ignored …
As originally demonstrated by McClintock, eukaryotic genome contain large number of transposable elements. In fact, at least 45 percent of the DNA in a human cell nucleus has been derived from transposable elements! The vast majority (>99 percent) of transposable elements are incapable of moving from one place to another, they have either been crippled by mutation or their movement is suppressed by the cell… However, when transposable elements do change potion, they insert widely throughout the target DNA. In fact, many transposable elements can insert themselves into the center of protein-coding gene. Several examples of such an occurrence have been documented in humans, including a number of cases of hemophilia caused by a mobile genetic element that had ‘jumped’ into the middle of the key blood clotting genes … In addition, the reactivation of transposable elements may contribute to the development of certain cancers.” (Gerald Karp, Cell Biology, p.143, Wiley)
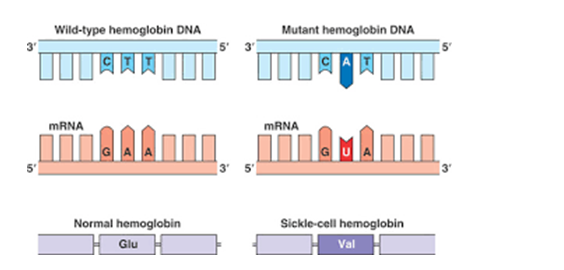
Fig. 13-2: The above shows a normal DNA segment on the left, and the same segment having received transposable element on the right. CTT (top-left) have been replaced by CAT-top right, and GAA (bottom-left) has been replaced by GUA and the result is appearance of Sickle-cell hemoglobin disease.
 Strangely however, such large occurrence of these transposable elements is not to be noticed in the animal kingdom. In contrast, their huge presence in the human genome is an unexplained phenomenon.
Strangely however, such large occurrence of these transposable elements is not to be noticed in the animal kingdom. In contrast, their huge presence in the human genome is an unexplained phenomenon.
Some of these transposable elements are in fact welcome by the cell. They can be beneficial. One of the functions is to adapt the body to newer physical environment. They influence gene expression by suppressing some or activating others.
Studies show that Transposition is just one of the many ways in which genome is altered. By virtue of this mechanism, apart from others, cells rearrange their own DNA. But the rearrangement is not haphazard. They follow certain precise rules. Otherwise, the body will die in minutes. Protozoans (a single-celled animal), are known to be able to rearrange – miraculously – 100,000 segments of their own DNA within a short time. And everything remains in order.
That is, reshuffling is not a ‘hit-and-miss’ venture. It is well directed and has fixed objectives. Not any segment of the DNA or a molecule can insert itself by its own will into the genome. The operation is not something like someone passing through a door to enter a room. There are no doors and no openings in the DNA strands. A transposable element has to cut open an entrance into the chromosome with the help of a machine. Nor can it be done just anywhere it wishes. There are marks at which alone the cutting through can be done. Further, the new transposable element needs to join its own two ends to the DNA chain. It’s an organized, deliberate process, after which predictable processes and consequences follow.
Figure below illustrates how a segment of DNA from a normal chromosome numbered 9, is translocated on chromosome numbered 22, resulting in a modified chromosome that results in diseases of leukemia.
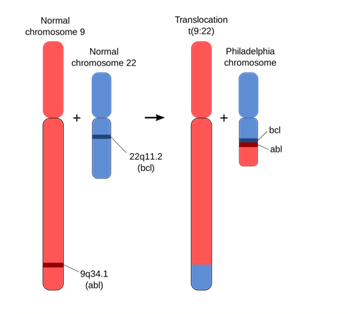
Philadelphia Chromosome
Fig. 13-3: An abnormality of chromosome 22 in which part of chromosome 9 is transferred to it. Bone marrow cells that contain the Philadelphia chromosome are often found in chronic myelogenous leukemia and sometimes found in acute lymphocytic leukemia.
There are of course several kinds of transposition elements, and their different ways of implanting themselves are pretty complicated. Our purpose is to point out that, human traits, or for that matter of any organism, are not entirely gene-dependent: such genes as have been inherited from the parents. There are factors that can modify the inherited genome. One of those factors is Transposition which plays an important role in the development of an organ or organism. In contrast, it is commonly asserted by the evolutionists that random mutation alone, given enough time, creates new traits and new species. Random mutation happens to be a pillar of the newest version of Neo-Darwinism, but which now faces collapse. A recent textual work in Biology notes:
“… transposition has had a profound impact on the genetic composition of organisms. It is interesting to note that, only a few decades ago, molecular biologists considered the genome to be a stable repository of genetic information. Now it seems remarkable that organisms can maintain themselves from one day to the next in the face of this large-scale disruption by genetic rearrangement.” (Cell Biology, Gerald Karp, Wiley, 2014)
Duplication
Another example of Transposition is duplication of genetic information. That is, insertion of genetic segments into chromosomes it doesn’t originally belong to.

Fig. 13-4: An illustration of how genetic duplication takes place.
We are aware that epigenetics or transposition elements and their roles are claimed by some biologists as in tandem with evolution. Yves Bigot wrote:
Sequencing various prokaryotic and eukaryotic genomes has revealed that transposable elements are ubiquitous among plants, bacteria, fungi and animals, and that they exhibit very diverse biological features. Their massive dominance in genomes means that they have been heavily involved in genome evolution, as they shape genome architecture and contribute to genetic innovations. (Mobile Genetic Elements, Edited by Yves Bigot, 2012, epub)
Our point however has been, as demonstrated by Perry Marshall in his work Evolution 2, that, in the light of these new elements, both the original as well as the modified version of the theory of evolution are now almost defunct. Those who are fighting for the life of the theory need to formulate a new version. For all we know, that’s not going to be easy. If attempted, it is bound to give birth to innumerable controversies – so advanced is the science of biology today and so massive is the information now available.
The entire emphasis in the past by the evolutionist has been on the role of point mutations in coding regions as the primary source of evolutionary change. On the other hand, the possibility that transposable elements, or other factors, could override genetic information, was, for the most part, ignored. Now it is being said that perhaps transposition is responsible in a large part for the appearance of new species. So be it. But the proof is yet to come.
The following may strain a non-biological reader, but proves our point that the phenomenon and mechanism known as Transposition, affects the DNA so much that the DNA of a biological organism, be it ducks, foxes or humans, is a helpless victim of influences coming from outside, and, in consequence, it is incorrect for the biologists to say that evolution is nothing but the result of “random mutations” working on the DNA, to change characteristics, and for “natural selection” to collect together the modifications and produce a new species.
The life-cycle of a TE [transposable element) family is akin to a birth-and-death process. The scenario is likely to proceed in three distinct phases. First, the initial phase of the TE invasion of a population does not generally proceed at a low and uniform rate, but tends to occur in sudden episodic bursts. If excessive disruption of a genome were to occur, this could lead to the extinction of the individual or lineage. This is the phase during which the genome is highly unstable and generates new combinations of genes and new regulatory networks. Second, after this invasion, there is a period of time during which the TE family proliferates within the population accompanied by mutations that render some elements inactive.
Most TE insertions are tolerated as a result of the repression of other members of the TE family; a kind of host defense mechanism. This period is a stage during which the genome is considered to be dynamic, with copy amplification and loss of copies roughly in balance. Finally, during a more quiescent state, transposition continues at a reduced rate, and selection against insertions is still required to stabilize the copy number. During this final phase, the TE could be extinguished as a result of the accumulation of mutations within TE copies, and some copies could be “recruited” by the genome for new functions through the domestication process. There are two main ways for TEs to escape extinction: by horizontal transfer to a new host genome prior to inactivation and by inflicting of minimal harmful effects by having a low replication rate so as to evade detection by the host. (Nonetheless) Little is known about the absolute rates of transposition because few of the elements described have been observed to be active in their host. (Mobile Genetic Elements, Edited by Yves Bigot, 2012, epub [internal references removed]).
(To be continued)

